Using OpenAI's GPT API models for Title and Abstract Screening in Systematic Reviews
Source:vignettes/Using-ChatGPT-For-Screening.Rmd
Using-ChatGPT-For-Screening.RmdImportant note
See Vembye et al. (2024) for an overview of how and when GPT API models can be used for title and abstract screening. For an overview of other research regarding the use of GPT API models for title and abstract screening, see Syriani et al. (2023), Guo et al., (2024), and Gargari et al. (2024). On a related line of research, Alshami et al. (2023), Khraisha et al. (2024), and Issaiy et al. (2024) explored the ChatGPT web browser interface for TAB screening. Based on our experience, we think that these two lines of research should not be mixed up since the draw on different GPT models.
In this vignette, we show how to conduct title and abstract screening
with OpenAI’s GPT API models in R. The advantages of conducting the
screening with ChatGPT via R is 1) that reviewers can easily work with a
large number of references, avoiding copy-paste procedures (cf. Khraisha
et al., 2024), 2) that the total screening time can be substantially
increased relative to using the ChatGPT interface, 3) that it eases
model comparison, and 4) that consistency between GPT answer for the
same title and abstract can easily be tested. In this vignette, we also
show the first/initial proof of concept for the use of GPT API models as
reliable second screeners (see also Syriani et al., 2023). This goes
without saying that the GPT API models work in all cases, and we think
the tool should be use carefully and should always assisted by a human
(human-in-the-loop). Consequently, we do not recommend to use the
AIscreenR as a single screener per se. If used to reduced
the overall number of references needed to be screened, our tentative
recommendation is to random sample approximately 10% of the studies
excluded by GPT to check the accuracy of the screening.
Getting started: Loading relevant ris file data for screening
At this stage, we expect that you have a pile of ris-files,
containing titles and abstracts for the references you would like to
screen. You can retrieve ris-files in several ways, either directly from
research database, a Google Scholar search, or exported from your
reference management tool, such as EndNote, Mendeley, and RefMan.
Alternatively, you can export ris-file from systematic software tool
such as EPPI-reviewer,
Covidence, MetaReviewer,
revtools, or whatever
software you use. In the example given below, we load ris-files
extracted from the EPPI-reviewer. A minor advantage of extracting
ris-files from systematic software tools is that they add a unique study
ID for each reference. This feature makes it easier to keep track of the
screening. Yet, such ID can be generated in R as well, and is
automatically generated in the tabscreen_gpt() function
when unique IDs are not provided. When using a GPT API model as the
second screener, we recommend that the human screening has been done
before uploading to R. Thereby, it is possible to compare the screenings
instantly after the computer screening has been done. In the below
example we load ris-files separately for excluded and included
ris-files, respectively, and add the human_code variable
that tracks the human decision.
# Installation
# install.packages("devtools")
# devtools::install_github("MikkelVembye/AIscreenR")
# Loading packages
library(AIscreenR)
library(revtools)
library(tibble)
library(dplyr)
library(purrr)
library(usethis)
library(future)
# Loading excluded studies
# Reading path to risfiles
excl_path <- system.file("extdata", "excl_tutorial.ris", package = "AIscreenR")
ris_dat_excl <- revtools::read_bibliography(excl_path) |>
suppressWarnings() |>
as_tibble() |>
select(author, eppi_id, title, abstract) |> # Using only relevant variables
mutate(
human_code = 0
)
# Loading included studies
incl_path <- system.file("extdata", "incl_tutorial.ris", package = "AIscreenR")
ris_dat_incl <- revtools::read_bibliography(incl_path) |>
suppressWarnings() |>
as_tibble() |>
select(author, eppi_id, title, abstract) |>
mutate(
human_code = 1
)
filges2015_dat <-
bind_rows(ris_dat_excl, ris_dat_incl) |>
mutate(
studyid = 1:n()
) |>
relocate(studyid, .after = eppi_id)
filges2015_dat
#> # A tibble: 270 × 6
#> author eppi_id studyid title abstract human_code
#> <chr> <chr> <int> <chr> <chr> <dbl>
#> 1 Holloway R G and Gramling R and Ke… 9434957 1 Esti… "Progno… 0
#> 2 Morawska Alina and Stallman Helen … 9433838 2 Self… "Behavi… 0
#> 3 Michel C M and Pascual-Marqui R D … 9431171 3 Freq… "The to… 0
#> 4 Paul Howard A 9433968 4 A Re… "The ar… 0
#> 5 Feinberg I and De Bie E and Davis … 9434460 5 Topo… "STUDY … 0
#> 6 Hamburg Sam R 9433554 6 BOOK… "The ar… 0
#> 7 Park H Y and Lee B J and Kim J H a… 9435130 7 Rapi… "Backgr… 0
#> 8 Petrek J 9432040 8 Pict… "AIMS: … 0
#> 9 Schwartzman Meredith P and Wahler … 9434093 9 Enha… "New an… 0
#> 10 Faber J and Srutova L and Pilarova… 9431505 10 EEG … "Sponta… 0Getting API key (OpenAI)
Before you can use the functions from AIscreenR to
screen your references, you must generate your own secret API key from
OpenAI. To do so you must first ensure that you have created an account
at OpenAI (if you have not done so at this stage, you can sign up here).
When having an account, go to https://platform.openai.com/account/api-keys
and press the + Create new secret key button (see Figure 1
below) and give your key a name.
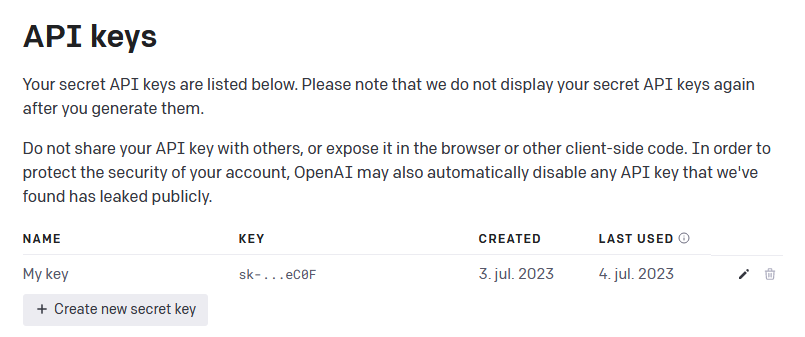
Figure 1 - Generate API key from OpenAI
When you have generated your secret API key, remember to store it safely since you will not be able see it again. NOTE: If you lose your API key, you can just generate a new one.
Handling your API key
When you have retrieved your API, you could in theory add it directly
to the AIscreenR functions via the api_key
argument. Yet, this would be an improper way to work the API key since
you would easily compromise your secret key. For example, your API key
would be disclosed when sharing your codes with others which in turn
would be able to draw on your OpenAI account. Also OpenAI will cancel
the API key if they recognize that your API key has been compromised,
e.g., by pushing it to a public GitHub page. To overcome this issue you
have several options. You can either work with what we call permanent or
temporary solutions.
Pemanent solution
The easiest way to work with your API key is to permanently added it
to your R environment as an environment variable. This can be achieved
with usethis::edit_r_environ(). In the
.Renviron file, write CHATGPT_KEY=your_key as
depicted in Figure 2. After entering the API key, close and save the
.Renviron file and restart RStudio (ctrl +
shift + F10). From now on, the AIscreenR functions will use
the get_api_key() function to get your API key from your R
environment automatically. By using this approach you don’t have to
worry more about you API key (unless you update RStudio, deliberately
delete the key, or get a new computer. Then you must repeat this
procedure).
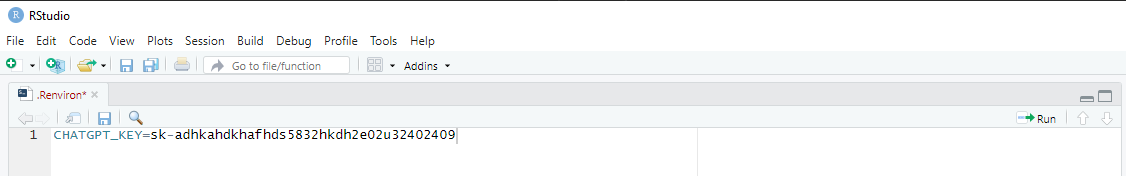
Figure 2 - R environment file
Temporary solution
If you do not want to add you API key permanently to your R
environment, you can use set_api_key(). When executing
set_api_key(), you will see a pop-up window in which you
can enter your API key. This will add your API key as temporary
environment variable. Consequently, when you restart RStudio, you will
no longer be able to find your API key in your R environment.
Alternatively, you can pass a decrypted key to the
set_api_key(), like
set_api_key(key = secret_decrypt(encrypt_key, "YOUR_SECRET_KEY_FOR_DECRYTING")).
See the HTTR2
pacakge for further details about this solution.
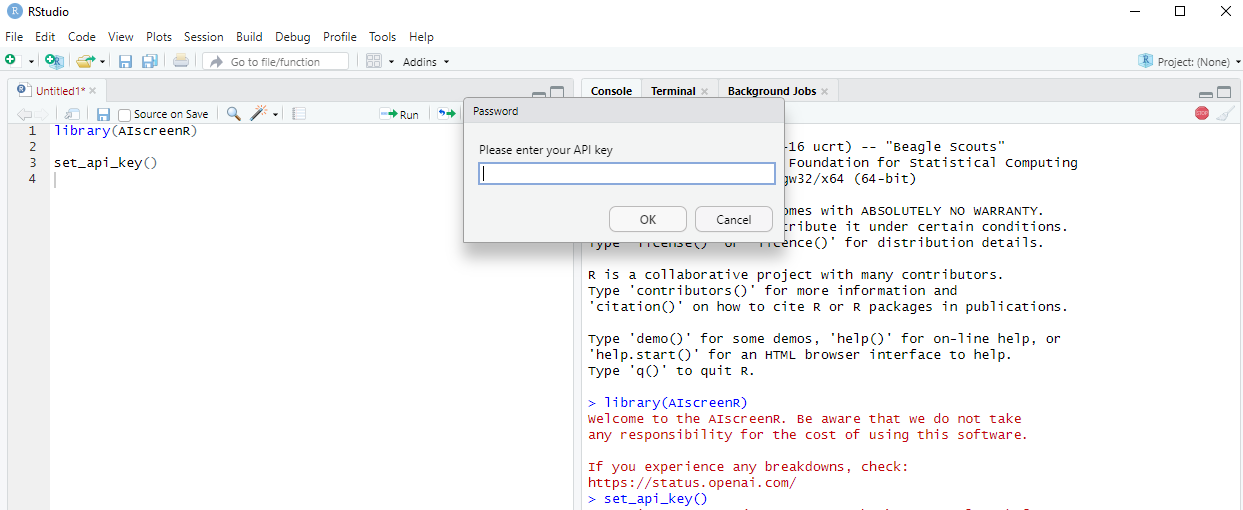
Figure 3 - Set API key
Working with prompts in R
Example of how to enter a prompt.
prompt <- "Evaluate the following study based on the selection criteria
for a systematic review on the effects of family-based interventions on drug
abuse reduction for young people in treatment for non-opioid drug use.
A family-based intervention (FFT) is equivalent to a behavior focused
family therapy, where young people’s drug use is understood in relation to
family behavior problems. Family-based interventions also includes manual-based
family therapies as it targets young people and their families as a system
throughout treatment, and thereby recognizes the important role of the family
system in the development and treatment of young people’s drug use problems.
FFT was developed in the late 1980s on request from the US National Institute on
Drug Abuse (NIDA). The development of FFT was initially heavily inspired by the
alcohol abuse program Community Reinforcement Approach (CRA), which was aimed
at restructuring the environment to reinforce non-alcohol associated activities.
FFT developed to have more emphasis on contingency contracting, impulse control
strategies specific to drug use, and increased emphasis on involvement of family
members in treatment. FFT is designed to accommodate diverse populations of
youths with a variety of behavioral, cultural and individual preferences.
FFT has evolved for use in severe behavioral disturbances known to co-exist with
substance use and dependence, and the core interventions have been enhanced to
address several mental health related problems commonly occurring
as comorbid conditions in drug use treatment participant. For each study,
I would like you to assess: 1) Is the study about a family-based intervention,
such as Functional Family Therapy, Multidimensional Family Therapy, or
Behavioral Family Therapy? (Outpatient manual-based interventions of any
duration delivered to young people and their families). If not, exclude study.
2) Are the participants in outpatient drug treatment primarily
for non-opioid drug use? 3) Are the participants within age 11–21?"Can also be done in word
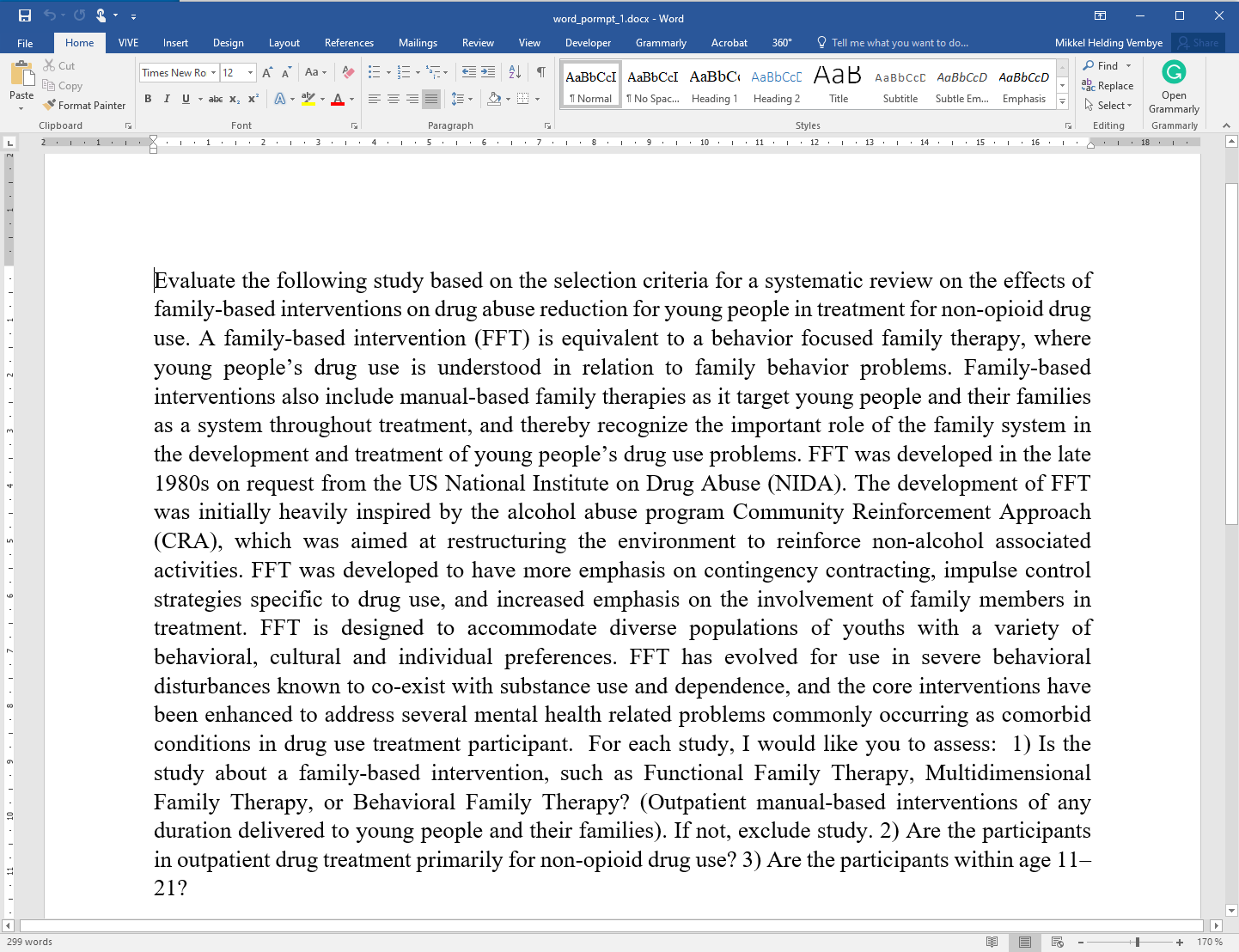
Figure 4 - Prompt 1: With three inclusion critera.
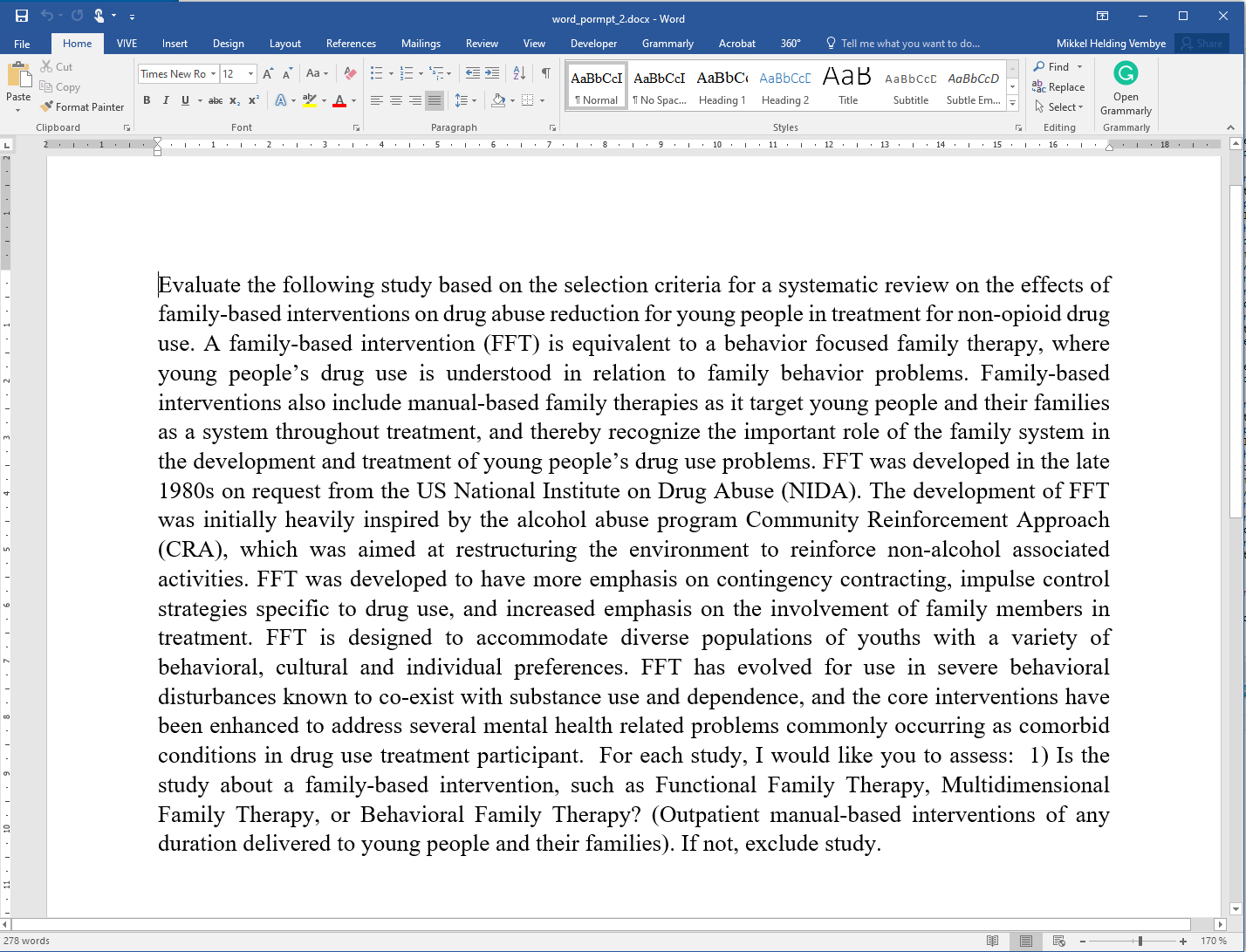
Figure 5 - Prompt 2: With one inclusion criterion only.
word_path <- system.file("extdata", "word_prompt_1.docx", package = "AIscreenR")
prompt1 <-
readtext::readtext(word_path)$text |>
stringr::str_remove_all("\n")
prompt1
#> [1] "Evaluate the following study based on the selection criteria for a systematic review on the effects of family-based interventions on drug abuse reduction for young people in treatment for non-opioid drug use. A family-based intervention (FFT) is equivalent to a behavior focused family therapy, where young people’s drug use is understood in relation to family behavior problems. Family-based interventions also include manual-based family therapies as it target young people and their families as a system throughout treatment, and thereby recognize the important role of the family system in the development and treatment of young people’s drug use problems. FFT was developed in the late 1980s on request from the US National Institute on Drug Abuse (NIDA). The development of FFT was initially heavily inspired by the alcohol abuse program Community Reinforcement Approach (CRA), which was aimed at restructuring the environment to reinforce non-alcohol associated activities. FFT was developed to have more emphasis on contingency contracting, impulse control strategies specific to drug use, and increased emphasis on the involvement of family members in treatment. FFT is designed to accommodate diverse populations of youths with a variety of behavioral, cultural and individual preferences. FFT has evolved for use in severe behavioral disturbances known to co-exist with substance use and dependence, and the core interventions have been enhanced to address several mental health related problems commonly occurring as comorbid conditions in drug use treatment participant. For each study, I would like you to assess: 1) Is the study about a family-based intervention, such as Functional Family Therapy, Multidimensional Family Therapy, or Behavioral Family Therapy? (Outpatient manual-based interventions of any duration delivered to young people and their families). If not, exclude study. 2) Are the participants in outpatient drug treatment primarily for non-opioid drug use? 3) Are the participants within age 11–21?"
# Working with multiple prompts
word_paths <- system.file("extdata", c("word_prompt_1.docx", "word_prompt_2.docx"), package = "AIscreenR")
prompts <-
purrr::map_chr(
word_paths, ~ {
readtext::readtext(.x)$text |>
stringr::str_remove_all("\n")
}
)
prompts
#> [1] "Evaluate the following study based on the selection criteria for a systematic review on the effects of family-based interventions on drug abuse reduction for young people in treatment for non-opioid drug use. A family-based intervention (FFT) is equivalent to a behavior focused family therapy, where young people’s drug use is understood in relation to family behavior problems. Family-based interventions also include manual-based family therapies as it target young people and their families as a system throughout treatment, and thereby recognize the important role of the family system in the development and treatment of young people’s drug use problems. FFT was developed in the late 1980s on request from the US National Institute on Drug Abuse (NIDA). The development of FFT was initially heavily inspired by the alcohol abuse program Community Reinforcement Approach (CRA), which was aimed at restructuring the environment to reinforce non-alcohol associated activities. FFT was developed to have more emphasis on contingency contracting, impulse control strategies specific to drug use, and increased emphasis on the involvement of family members in treatment. FFT is designed to accommodate diverse populations of youths with a variety of behavioral, cultural and individual preferences. FFT has evolved for use in severe behavioral disturbances known to co-exist with substance use and dependence, and the core interventions have been enhanced to address several mental health related problems commonly occurring as comorbid conditions in drug use treatment participant. For each study, I would like you to assess: 1) Is the study about a family-based intervention, such as Functional Family Therapy, Multidimensional Family Therapy, or Behavioral Family Therapy? (Outpatient manual-based interventions of any duration delivered to young people and their families). If not, exclude study. 2) Are the participants in outpatient drug treatment primarily for non-opioid drug use? 3) Are the participants within age 11–21?"
#> [2] "Evaluate the following study based on the selection criteria for a systematic review on the effects of family-based interventions on drug abuse reduction for young people in treatment for non-opioid drug use. A family-based intervention (FFT) is equivalent to a behavior focused family therapy, where young people’s drug use is understood in relation to family behavior problems. Family-based interventions also include manual-based family therapies as it target young people and their families as a system throughout treatment, and thereby recognize the important role of the family system in the development and treatment of young people’s drug use problems. FFT was developed in the late 1980s on request from the US National Institute on Drug Abuse (NIDA). The development of FFT was initially heavily inspired by the alcohol abuse program Community Reinforcement Approach (CRA), which was aimed at restructuring the environment to reinforce non-alcohol associated activities. FFT was developed to have more emphasis on contingency contracting, impulse control strategies specific to drug use, and increased emphasis on the involvement of family members in treatment. FFT is designed to accommodate diverse populations of youths with a variety of behavioral, cultural and individual preferences. FFT has evolved for use in severe behavioral disturbances known to co-exist with substance use and dependence, and the core interventions have been enhanced to address several mental health related problems commonly occurring as comorbid conditions in drug use treatment participant. For each study, I would like you to assess: 1) Is the study about a family-based intervention, such as Functional Family Therapy, Multidimensional Family Therapy, or Behavioral Family Therapy? (Outpatient manual-based interventions of any duration delivered to young people and their families). If not, exclude study. "Retrieve rate limit information
# Rate limits across one model (Default is "gpt-3.5-turbo-0613")
rate_limits <- rate_limits_per_minute()
rate_limits
#> # A tibble: 1 × 3
#> model requests_per_minute tokens_per_minute
#> <chr> <dbl> <dbl>
#> 1 gpt-3.5-turbo-0613 10000 1000000
# Rate limits overview across multiple models
# Add further models if necessary
models <- c("gpt-3.5-turbo-0613", "gpt-4")
models_rate_limits <- rate_limits_per_minute(model = models)
models_rate_limits
#> # A tibble: 2 × 3
#> model requests_per_minute tokens_per_minute
#> <chr> <dbl> <dbl>
#> 1 gpt-3.5-turbo-0613 10000 1000000
#> 2 gpt-4 200 10000Approximate price of screening
Approximate price of screening
app_obj <-
approximate_price_gpt(
data = filges2015_dat, # Tutorial data embedded in the package
prompt = prompts,
studyid = studyid, # indicate the variable with the studyid in the data
title = title, # indicate the variable with the titles in the data
abstract = abstract, # indicate the variable with the abstracts in the data
model = c("gpt-3.5-turbo-0613", "gpt-3.5-turbo-0613", "gpt-4"),
reps = c(1, 10, 1),
top_p = c(0.001, 1)
)
app_obj
#> The approximate price of the (simple) screening will be around $56.1396.
app_obj$price_dollar
#> [1] 56.1396
app_obj$price_data
#> # A tibble: 6 × 6
#> prompt model iterations input_price_dollar output_price_dollar
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Prompt 1 gpt-3.5-turbo-0613 1 0.9034 0.01188
#> 2 Prompt 1 gpt-3.5-turbo-0613 10 9.042 0.1188
#> 3 Prompt 1 gpt-4 1 18.08 0.3564
#> 4 Prompt 2 gpt-3.5-turbo-0613 1 0.875 0.01188
#> 5 Prompt 2 gpt-3.5-turbo-0613 10 8.755 0.1188
#> 6 Prompt 2 gpt-4 1 17.51 0.3564
#> total_price_dollar
#> <dbl>
#> 1 0.9153
#> 2 9.161
#> 3 18.44
#> 4 0.8869
#> 5 8.874
#> 6 17.87Screening titles and abstracts
models <- c("gpt-3.5-turbo-0613", "gpt-3.5-turbo-0613", "gpt-4")
reps <- c(1, 10, 1)
rpm <- c(10000, 10000, 200)
plan(multisession)
result_object <-
tabscreen_gpt(
data = filges2015_dat, # ris-file data create above
prompt = prompt, # indicate name of the loaded prompt object
studyid = studyid, # indicate the variable with the studyid in the data
title = title, # indicate the variable with the titles in the data
abstract = abstract, # indicate the variable with the abstracts in the data,
model = c("gpt-3.5-turbo-0613", "gpt-3.5-turbo-0613", "gpt-4"),
reps = c(1, 10, 1),
rpm = c(10000, 10000, 200),
top_p = c(0.001, 1)
)
#> * The approximate price of the current (simple) screening will be around $27.9901.
#> * Consider removing references that has no abstract since these can distort the accuracy of the screening
#> Progress: ──────────────────────────────────────────────────────────────────────────────────────────── 100%
#> * NOTE: Requests failed 4 times."
plan(sequential)
print(result_object)
#> Find data with all answers by executing
#> result_object$answer_data_all
#>
#> Find data with the result aggregated across multiple answers by executing
#> result_object$answer_data_sum
#>
#> Find total price for the screening by executing
#> result_object$price_dollar
#>
#> Find error data by executing
#> result_object$error_dataScreen failed requests
result_object <-
result_object |>
screen_errors()
print(result_object)
#> Find data with all answers by executing
#> result_object$answer_data_all
#>
#> Find data with the result aggregated across multiple answers by executing
#> result_object$answer_data_sum
#>
#> Find total price for the screening by executing
#> result_object$price_dollarGetting the results from the screening
# gpt-3.5 results (prompt 1, top_p = 1, 10 replications)
# studyid = 21 is not included since it failed during the loop
# I show how to recover this in another
sum_dat_gpt3 <-
result_object$answer_data_sum |>
filter(stringr::str_detect(model, "3") & promptid == 1 & reps == 10 & top_p == 1)
# gpt-4 results (prompt 1, top_p = 1)
sum_dat_gpt4 <-
result_object$answer_data_sum |>
filter(stringr::str_detect(model, "4") & promptid == 1 & top_p == 1)Analyzing the screening
False included and excluded (by gpt)
screen_perform <-
result_object |>
screen_analyzer()
screen_perform
#> # A tibble: 12 × 25
#> promptid model reps top_p n_refs n_screened n_missing human_in_gpt_ex
#> <int> <chr> <int> <dbl> <int> <int> <int> <dbl>
#> 1 1 gpt-3.5-tur… 1 0.001 135 135 0 8
#> 2 1 gpt-3.5-tur… 10 0.001 135 135 0 8
#> 3 1 gpt-3.5-tur… 1 1 135 135 0 11
#> 4 1 gpt-3.5-tur… 10 1 135 135 0 4
#> 5 1 gpt-4 1 0.001 135 135 0 1
#> 6 1 gpt-4 1 1 135 135 0 0
#> 7 2 gpt-3.5-tur… 1 0.001 135 135 0 9
#> 8 2 gpt-3.5-tur… 10 0.001 135 135 0 8
#> 9 2 gpt-3.5-tur… 1 1 135 135 0 10
#> 10 2 gpt-3.5-tur… 10 1 135 135 0 7
#> 11 2 gpt-4 1 0.001 135 135 0 1
#> 12 2 gpt-4 1 1 135 135 0 2
#> human_ex_gpt_in human_in_gpt_in human_ex_gpt_ex accuracy p_agreement
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 8 37 82 0.1185 0.8815
#> 2 8 37 82 0.1185 0.8815
#> 3 8 34 82 0.1407 0.8593
#> 4 8 41 82 0.08889 0.9111
#> 5 9 44 81 0.07407 0.9259
#> 6 6 45 84 0.04444 0.9556
#> 7 6 36 84 0.1111 0.8889
#> 8 7 37 83 0.1111 0.8889
#> 9 6 35 84 0.1185 0.8815
#> 10 7 38 83 0.1037 0.8963
#> 11 8 44 82 0.06667 0.9333
#> 12 7 43 83 0.06667 0.9333
#> precision recall npv specificity bacc F2 mcc irr se_irr
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.8222 0.8222 0.9111 0.9111 0.8667 0.5139 0.8667 0.7333 0.01236
#> 2 0.8222 0.8222 0.9111 0.9111 0.8667 0.5139 0.8667 0.7333 0.01236
#> 3 0.8095 0.7556 0.8817 0.9111 0.8333 0.4885 0.8394 0.6780 0.01308
#> 4 0.8367 0.9111 0.9535 0.9111 0.9111 0.5452 0.9030 0.8043 0.01113
#> 5 0.8302 0.9778 0.9878 0.9 0.9389 0.5612 0.9237 0.8404 0.01046
#> 6 0.8824 1 1 0.9333 0.9667 0.5859 0.9537 0.9032 0.008146
#> 7 0.8571 0.8 0.9032 0.9333 0.8667 0.5172 0.8734 0.7458 0.01182
#> 8 0.8409 0.8222 0.9121 0.9222 0.8722 0.5197 0.8744 0.7486 0.01195
#> 9 0.8537 0.7778 0.8936 0.9333 0.8556 0.5087 0.8645 0.7273 0.01209
#> 10 0.8444 0.8444 0.9222 0.9222 0.8833 0.5278 0.8833 0.7667 0.01166
#> 11 0.8462 0.9778 0.9880 0.9111 0.9444 0.5670 0.9305 0.8556 0.009913
#> 12 0.86 0.9556 0.9765 0.9222 0.9389 0.5658 0.9284 0.8541 0.009807
#> # ℹ 3 more variables: cl_irr <dbl>, cu_irr <dbl>, level_of_agreement <chr>References
Alshami, A., Elsayed, M., Ali, E., Eltoukhy, A. E. E., & Zayed, T. (2023). Harnessing the power of ChatGPT for automating systematic review process: Methodology, case study, limitations, and future directions. Systems. 11(7).
Gargari, O. K., Mahmoudi, M. H., Hajisafarali, M., & Samiee, R. (2024). Enhancing title and abstract screening for systematic reviews with GPT-3.5 turbo. BMJ Evidence-Based Medicine, 29(1), 69 LP – 70. https://doi.org/10.1136/bmjebm-2023-112678
Guo, E., Gupta, M., Deng, J., Park, Y.-J., Paget, M., & Naugler, C. (2024). Automated Paper Screening for Clinical Reviews Using Large Language Models: Data Analysis Study. Journal of Medical Internet Research. https://doi.org/10.2196/48996
Issaiy, M., Ghanaati, H., Kolahi, S., Shakiba, M., Jalali, A. H., Zarei, D., Kazemian, S., Avanaki, M. A., & Firouznia, K. (2024). Methodological insights into ChatGPT’s screening performance in systematic reviews. BMC Medical Research Methodology, 24(1), 78. https://doi.org/10.1186/s12874-024-02203-8
Khraisha Q, Put S, Kappenberg J, Warraitch A, Hadfield K (2024). Can large language models replace humans in systematic reviews? Evaluating GPT-4’s efficacy in screening and extracting data from peer-reviewed and grey literature in multiple languages. Research Synthesis Methods. 1-11. https://doi.org/10.1002/jrsm.1715
Syriani, E., David, I., & Kumar, G. (2023). Assessing the Ability of ChatGPT to Screen Articles for Systematic Reviews. ArXiv Preprint ArXiv:2307.06464. https://arxiv.org/pdf/2307.06464.pdf
Vembye, M., Christensen, J., Mølgaard, A. B., Schytt, F.L.W. (2024). GPT API Models Can Function as Highly Reliable Second Screeners of Titles and Abstracts in Systematic Reviews: A Proof of Concept and Common Guidelines. Open Science Framework (OSF). https://doi.org/10.31219/osf.io/yrhzm
Westgate MJ (2019). revtools: An R package to support article screening for evidence synthesis. Research Synthesis Methods. https://doi.org/10.1002/jrsm.1374.